We present here the anwser to some questions that are more common to
arise while running ENMTML. If you couldn’t find your issue
among those questions, please report it and we
will make our best to help you run the package smoothly.
At first the number fo arguments and the folder’s structure may seem a
little scary, but with little time you will get the hang of it and will
start running ENM models!
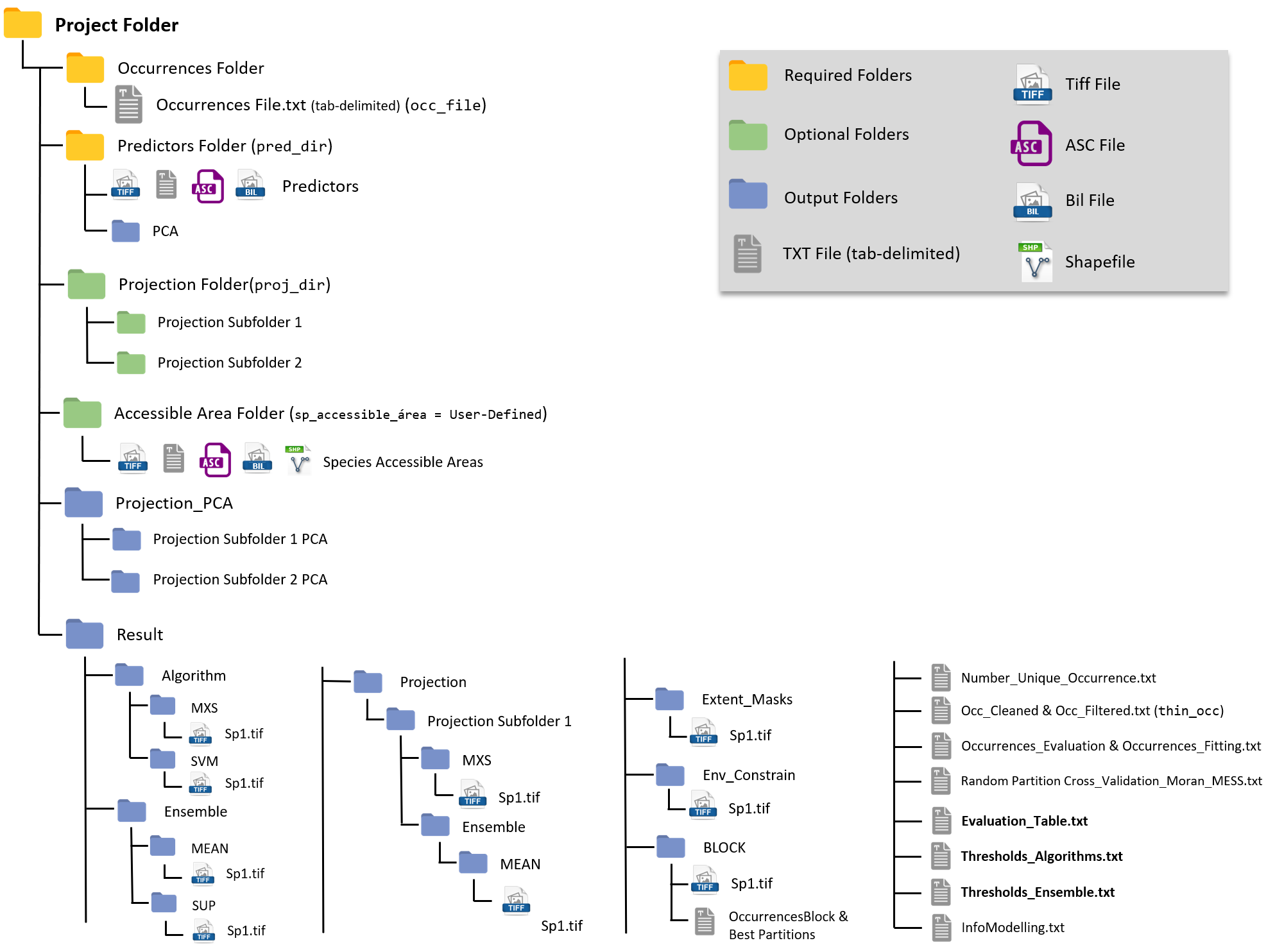
Project Folder
We highly recommend you to create a folder for your
project!
A general overview of the Project Folder and its containing sub-folders
will look similar to this:
Yellow are the Mandatory Folders, you need
them to run the package.
Green are the Optional Folders, only required according to your
project design.
Blue are Output Folders, those will be created during the
modeling routine.
Function Arguments
ENMTML consists of several arguments, which will be
checked at the start of the modeling routine.
Take your time to check any Error returned during this
argument check and use the help to correctly fill the
arguments.
Occurrence Data
Occurrence File Type
ENMTML imports occurrence data from a
TXT file with tabulation as separator
(tab-separated)
Data Format
The TXT can have several columns, but three columns are mandatory:
- Species name
- Longitude
- Latitude
They can be named to your preference as those will be inputted in the
arguments sp, x, y of the ENMTML
function.
Here’s an example of an occurrence table for four sample species.
#> Loading required package: ENMTML
#> Please note that rgdal will be retired during October 2023,
#> plan transition to sf/stars/terra functions using GDAL and PROJ
#> at your earliest convenience.
#> See https://r-spatial.org/r/2023/05/15/evolution4.html and https://github.com/r-spatial/evolution
#> rgdal: version: 1.6-7, (SVN revision 1203)
#> Geospatial Data Abstraction Library extensions to R successfully loaded
#> Loaded GDAL runtime: GDAL 3.6.2, released 2023/01/02
#> Path to GDAL shared files: C:/Users/santi/AppData/Local/R/win-library/4.3/rgdal/gdal
#> GDAL does not use iconv for recoding strings.
#> GDAL binary built with GEOS: TRUE
#> Loaded PROJ runtime: Rel. 9.2.0, March 1st, 2023, [PJ_VERSION: 920]
#> Path to PROJ shared files: C:/Users/santi/AppData/Local/R/win-library/4.3/rgdal/proj
#> PROJ CDN enabled: FALSE
#> Linking to sp version:2.1-1
#> To mute warnings of possible GDAL/OSR exportToProj4() degradation,
#> use options("rgdal_show_exportToProj4_warnings"="none") before loading sp or rgdal.
#> Registered S3 methods overwritten by 'adehabitatMA':
#> method from
#> print.SpatialPixelsDataFrame sp
#> print.SpatialPixels sp
#> rgeos version: 0.6-4, (SVN revision 699)
#> GEOS runtime version: 3.11.2-CAPI-1.17.2
#> Please note that rgeos will be retired during October 2023,
#> plan transition to sf or terra functions using GEOS at your earliest convenience.
#> See https://r-spatial.org/r/2023/05/15/evolution4.html for details.
#> GEOS using OverlayNG
#> Linking to sp version: 2.1-1
#> Polygon checking: TRUE| species | x | y |
|---|---|---|
| Sp_14 | -55.37526 | -31.708447 |
| Sp_14 | -54.20860 | -25.041806 |
| Sp_14 | -58.29191 | -29.125124 |
| Sp_17 | -73.12519 | -15.041847 |
| Sp_17 | -70.87520 | -15.458512 |
| Sp_17 | -69.12520 | -17.708503 |
| Sp_18 | -61.37523 | -26.625134 |
| Sp_18 | -61.29190 | -26.958465 |
| Sp_18 | -61.37523 | -23.125148 |
| Sp_34 | -67.29188 | -5.041887 |
| Sp_34 | -71.20853 | -7.458544 |
| Sp_34 | -68.20854 | -7.208545 |
Predictors
Predictor File Type
ENMTML accepts predictors from several formats:
TIF, BIL, ASC and
TXT(tab-separated)
Folder Arrangement
Predictors must be within a single folder, which
will be informed through the pred_dir argument.
Projections
Projections are indicated using the proj_dir
argument.
As it is common to usually make projections to multiple climate
scenarios or extents, the arrangement of Projection folders
consists in a major folder with different sub-folders (one for each
climatic scenario or extent).
It is essential that the name of the variables in those folder
match the names of variables within the pred_dir
folder!
Results
Results will be in a folder specified in the result_dir
argument, or if you chose not to supply any folder ENMTML
will create a folder named Results at the same level of your
pred_dir folder (see this at the figure at the Project Folder section.
Model Evaluation and information about the modelling routine can be
found in several txt files within the Results folder.
Important sub-folders with result maps are:
- Algorithm: with tif present projections for
the selected algorithms and the binary folders for the selected
thresholds
- Ensemble: with tif projections for the
selected ensembles.
- Projection: with tif projections for the
different climatic scenarios/extents
Other sub-folders will be created according to the modeling routine and contain masks related to pseudo-absence allocation and geographical partition of datasets.